Abstract
A comparison was made of 37 STR markers on the Y-chromosome of eight individuals bearing the surname Briese. Based on paper records, two of these individuals were known to be related, but relationships could not be determined for others. The genetic study showed three distinct genetic lineages bearing the Briese surname. Five of the eight were shown to have had a common ancestor within genealogical time, which suggests that a cluster of related Briese families lived in the Deutsch Krone area of West Prussia in the 18th century. A sixth individual, originating from a different geographic region, Meseritz in Posen, was more distantly related, while the remaining two individuals were not related at all, either to the group or to each other. While not providing proof, these findings agree with the hypothesis that the Briese surname originated as a place name that was initially adopted by unrelated people. This would result in distinct clusters of more-closely related families, genetically distinct from other such clusters. Genetic genealogy can help to determine relationships between emigrant Briese families and those remaining in Germany. More individuals from Briese families with known origins are needed to participate in this study to confirm and enlarge on these preliminary findings.
Introduction
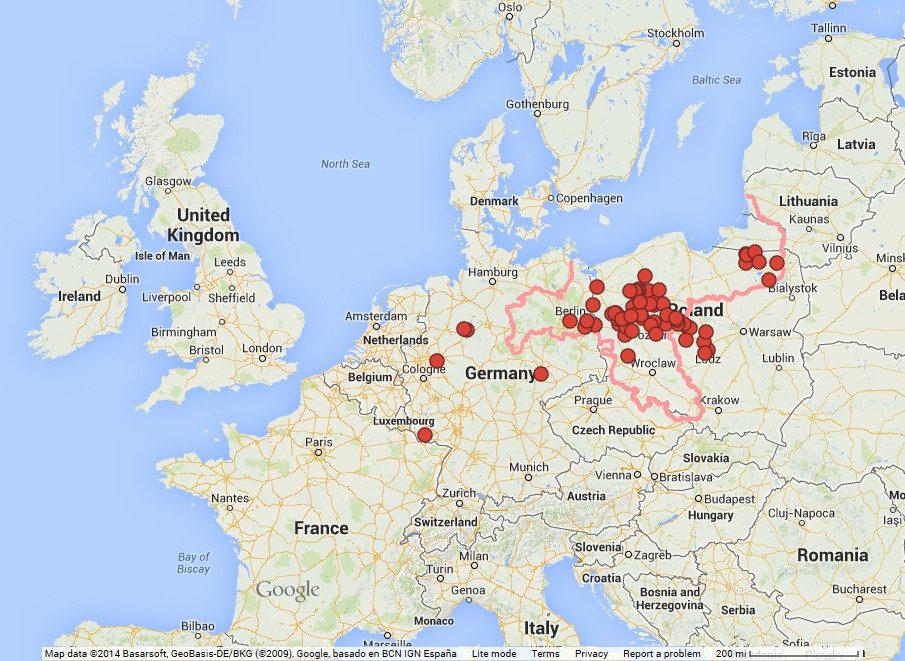
The Briese surname1 , as considered here, is of German origin. It is a place name, that is when people began adopting surnames in the middle ages, they took the name of the place where they lived. It is also a relatively uncommon surname, with a frequency of 30 per million in Germany and less in parts of the world to where Brieses have emigrated17 . More detail of the origins of the name can be found elsewhere1 . The distribution of the Briese surname during the 17th to 19th centuries is shown in Fig. 1. It indicates that the Briese heartland lies in the former Prussian provinces of Brandenburg, West Prussia and Posen, with a pocket occurring in East Prussia and a scattering in the west of Germany. This heartland area has many towns and villages bearing the name Briese, Briesen, or variants of this, and one or more of these are likely sources of the surname.
Fig. 1: Distribution of the Briese surname
Locations where the Briese surname appears in birth, death and marriage records from the 18th and early 19th century23415. The borders of the former eastern part of Prussia are shown in pink.
Being a place name suggests multiple origins, where unrelated people would have initially adopted the name Briese, although a single-source origin cannot be ruled out. In both cases, this would lead to clusters of related family groups, as families grew and moved. Classical genealogical research indicates that such geographic Briese clusters existed in the late 18th and 19th centuries in Brandenburg, West Prussia, East Prussia, Pomerania and Posen234 (see Table 1). However, subsequent internal movement, emigration overseas and displacement due to war has destroyed much of this demographic pattern. If the surname has multiple origins, some of these clusters should be differentiated from other such clusters, a possibility that is testable by genetic comparisons.
During the 19th century, a number of Briese families emigrated from these areas to North America, Australia and Brazil5. Many of the descendants of these families are now interested in discovering their family roots and whether their Briese family is related to other Briese families. Paper records provide some answers, but in many cases these are missing, have critical gaps, or do not go back far enough to provide the evidence needed. As both the surname and the Y-chromosome are passed down the male lineage of a family, genetic genealogy provides a complementary approach that can help unravel the answers to these questions.
This paper describes a study,set up in 20116 to describe the phylogenetic relationships between people bearing the surname Briese, based on the comparison of genetic markers on male Y-DNA. As well as determining patterns of family relationship, the results of such a study could also clarify conjecture on the origins of the Briese name.
Regions of Prussia with relatively high densities of the Briese name in 18th and early 19th century records.
Province
District
Brandenburg
Landsberg /Warthe
Brandenburg
Lebus
East Prussia
Gerdauern
Greater Poland
Lodz
Posen
Meseritz
Posen
Czarnikau
Posen
Schwerin
West Prussia
Deutsch Krone
West Prussia
Flatow
Methods
Raw data
To date, eight individuals with the surname Briese have contributed Y-DNA samples for analysis. These individuals descend from families that migrated to North America and Australia and, in all but two cases, can trace their family back to a specific location in Prussia. Genetic analyses were carried out by FTDNA (Family Tree DNA)7 . The recommended test for 37 STR (short tandem repeat) markers on the Y-chromosome was used, as the 12- and 25-marker sets offered are insufficient to determine relationships as accurately as required.
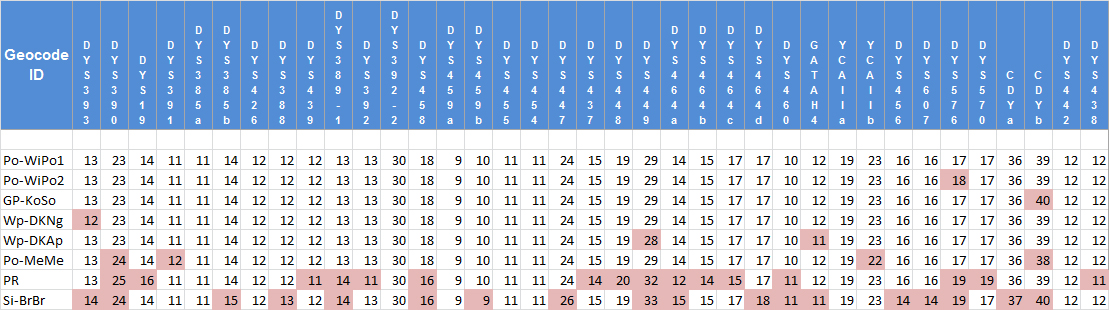
The data set of 37 STR markers is shown in Fig. 2. Individuals participating in this study are identified here by a 6-letter code to protect privacy. The code also identifies the location of the place where the earliest known ancestor of each individual lived (see Table 2).
Fig. 2: The Y-STR data set
De-identified data from the FamilyTreeDNA analysis of 37 STR markers on the Y-chromosome (coloured values show mutations from the modal haplotype). Original data are held at the Briese Surname Project6 .
Explanation of the geographic identity code (all regions are former parts of Prussia or neighbouring areas). A final number distinguishes individuals originating from the same location, where required e.g. Po-WiPo1 = Posen, Witkowo, Potrzymowo, person 1.
First letter
Next two letters
Last two letters
=Province (Provinz)
= District (Kreis)
= town or village
Po
Posen
Me
Meseritz
Me
Meseritz
Wi
Witkowo
Po
Potrzymowo
Wp
Westpreussen
DK
Deutsch Krone
Ap
Appelwerder
Ng
Neugolz
GP
Greater Poland
Ko
Konin
So
Sompolno
Si
Silesia
Br
Breslau
Br
Breslau
PR
Prussia (region unknown)
Genetic distances and phylogenetic tree
McGee’s Y-DNA Comparison Utility8 was used to produce a matrix of genetic distances (the total number of differences, or mutations, between two sets of results) based on the hybrid model used by FTDNA to calculate genetic mutations. This uses the stepwise mutation model for all alleles except DYS464 and YCAII, which use the infinite allele model8 . Using a mutation rate of 0.054 per generation (the average value derived from all FTDNA data), these data were then used to estimate the time to the most recent common ancestor (TMRCA) in numbers of generations. Individuals Po-WiPo1 and Po-WiPo2 are known to be related with 4 and 5 generations separation, respectively, from their most recent common ancestor and this was used to further calibrate the results.
TMRCAs were further analysed by the Kitsch program of Phylip10 (version 3.69), a freeware package of phylogenetic software. This program used the Fitch-Margoliash distance matrix method (with molecular clock) to generate the tree structure, showing the phylogenetic relationships of the eight individuals and, by inference, their families. The phylogenetic tree was drawn from this data using the open source genetic analysis package Mega611.
A time-line for the phylogenetic tree was then estimated, based on a generation time of 34 years (the median paternal generation interval of the project participants, going back to their most distant known ancestor (n=26)). This enabled the tree to be shown on two scales, the indicative number of generations back to the point when family lineages separated, and a calendar year time-line, which indicates when such separation may have occurred.
Network Analysis
Network analysis provides a useful means of visualising the pattern of DNA mutations between individuals or groups. McGee’s on-line Y-DNA Comparison Utility
Results
Genetic distances and the phylogenetic tree
Seven of the eight individuals belong to haplogroup r1b1 and the other (PR) to haplogroup r1a (Fig. 1), indicating a long genetic separation between them. The matrix of genetic distances between pairs of Briese individual haplotypes (Table 3) confirms this and refines the relationships between the group of seven. According to the FTDNA interpretation of genetic distance13, a distance of 1 indicates a tight relationship, 2-3 indicates that they are related, 4 indicates a probable relationship, 5 a possible relationship and 6 or more suggests relationships may pre-date the time when people in Europe adopted surnames.
Given that Po-WiPo1 and Po-WiPo2 are from the same family originating from Potrzymowo, Posen, it is not surprising that the genetic distance is so close (=1). This family has been in Australia since 1855. However, these two are also genetically close (distances = 2-3) to the two individuals originating from Deutsch Krone, Westpreussen (Wp-DKNg and Wp-DKAp). The family of Wp-DKNg emigrated to Australia and the family of Wp-DKAp emigrated to the USA. No connection had been found between these three families in paper records (births, deaths and marriages), but the y-DNA evidence demonstrates that they have a relatively recent common origin. The times to the most recent common ancestor of pairs of individuals indicate that were part of a common family about seven generations ago. A fourth family line, GP-KoSo which emigrated from nearby Sompolno to the United States, also forms part of this related cluster (distances = 1-3).
Matrix of genetic distances between pairs of Briese individuals (1-3 = related, 4-5 = probably related)
ID
Po-WiPo1
Po-WiPo2
Po-KoSo
Wp-DKNg
Wp-DKAp
Po-MeMe
PR
Si-BrBr
Po-WiPo1
1
1
1
2
4
23
27
Po-WiPo2
2
2
3
5
22
26
Po-KoSo
2
3
4
24
26
Wp-DKNg
3
5
24
28
Wp-DKAp
6
25
27
Po-MeMe
25
29
PR
27
Si-BrBr
Po-MeMe, originating from Meseritz, Posen, and now based in the USA, is genetically more separated from this group (distances = 4-6) and, when individual pairs are considered, borders on the limit of what FTDNA considers evidence for a relationship. However, the results need to be interpreted in terms of the overall pattern of genetic distances (see Fig. 3). The phylogenetic tree in Fig. 4 suggests that the split between the Meseritz family and the others occurred around 15 generations ago, i.e. in the early 16th century.
The last two individuals are clearly unrelated to the other six or to each other in genealogical time, showing very large genetic separation from them (distances = 22-29). As mentioned above, one even belongs to a different haplogroup (R1a) than members of the cluster (R1b). One of these is from a family that emigrated to the USA in the 19th century, but of unknown origin (other than Prussia). The other is from a family that emigrated from Silesia to the USA in the mid-20th century and whose oral tradition is of a Prussian origin.
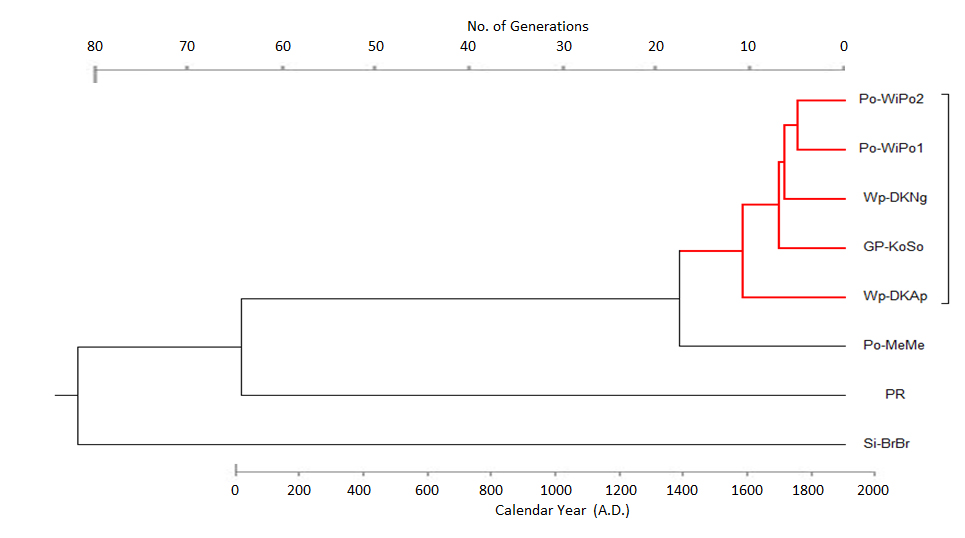
Fig. 3: Phylogenetic relationships of eight Briese individuals from seven separate “families”.
Each individual is identified by a code indicating the place where their earliest known ancestor was born or lived (see methods). The top axis shows the number of generations before now and the bottom axis is the calendar year (based on a 34-year generation interval).
Network Analysis
The results of the network analysis are shown in Fig. 3, which includes the whole network generated for the eight individuals and an enlargement of the nodal area. The network indicates that the unrelated individuals (PR and Si-BrBr) are separated from a grouping formed by the remaining six. The pattern of this cluster (see enlargement) is suggestive that there could be further sub-division.
Po-WiPo1 has a STR marker set that is identical to the modal haplotype. He is also known to be related to Po-WiPo2 from paper records, who together with GP-KoSo, Wp-DKNg and Wp-DKAp all lie within 1-2 mutations of the modal haplotype. These form a putative grouping of more closely related individuals and define the “Deutsch Krone” cluster, based on a region in West Prussia which in the 18th-19th centuries had a relatively high density of Briese surnames12
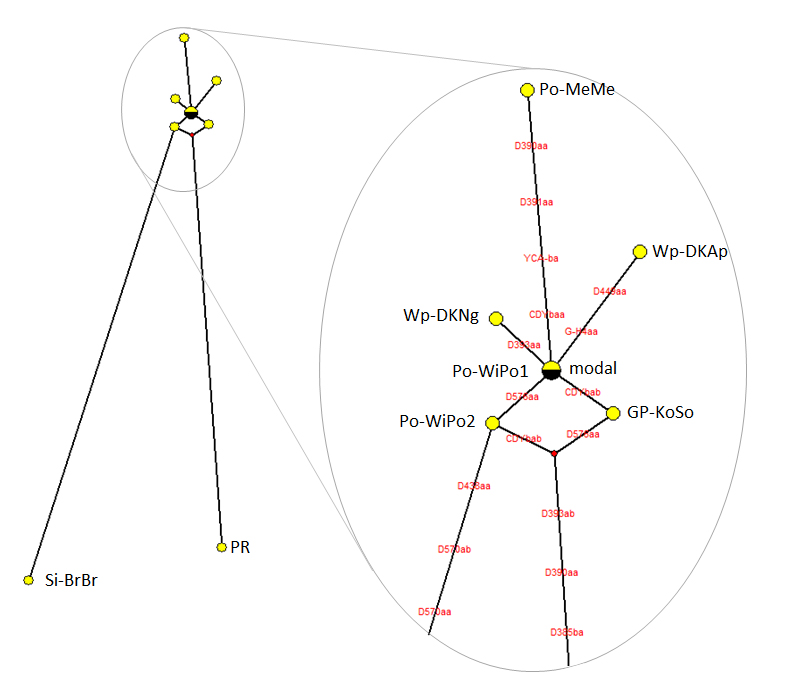
Fig. 4: Phylogenetic network for eight individuals with the surname Briese
The left-hand figure shows the full network and the right-hand figure is and enlargement of the upper cluster. The modal haplotype is shown in black and mutations from the modal haplotype are indicated in red (see text for details).
Discussion
A sample size of eight is small and, as a result, findings should be viewed as indicative. That said, the results have shown that genetic studies can make an important contribution to genealogy. They show that there are currently three distinct and unrelated lineages amongst people bearing the Briese surname.
Five of the individuals, two from Potrzymowo, Posen, two from Deutsch Krone, West Prussia (from the villages of Appelwerder and Neugolz, respectively) and one from Sompolno in Greater Poland, belong to a related cluster of Brieses, which was not demonstrable from paper records. The validity of this cluster is supported by two different lines of evidence. In the FTDNA y-search database18 of over 139,000 records, only one other person of different surname could possibly have been related (genetic distances of 3-5) at the 37-marker level, and this was subsequently disproved by a 67-marker STR comparison. Moreover, Immel et al. (2009)19found a high degree of correlation between surname and Y-STR haplotype in the East German population, suggesting that “Y-chromosomal genetic structure of modern Central European populations is heterogeneous and that, particularly in East Germany, the concomitant strata may be resolvable by the consideration of surnames”19 . Thus the close genetic relationships of these Briese individuals are highly unlikely to be due to chance.
The Y-DNA data suggest that the common ancestor lived in the late 17th or early 18th century. As mentioned previously, church records from the Lüben Evangelical Parish14 show that there were a large number of Brieses in the Deutsch Krone district, as far back as the mid 18th century. By contrast, the related Briese family from Potrzymowo is the only one in the Czerniejewo Evangelical Church Records15. This suggests that the family may have moved to that area from Deutsch Krone just prior to the dates of surviving paper records. With local movements such as these, geography alone does not define a family cluster.
The individual originating from Meseritz also appears to be related to the Deutsch Krone cluster, but more distantly, dating back perhaps to the 15th century. Given the large number of Briese records in the Meseritz church book215 in the 18th and 19th century, it seems likely that this region comprises a distinct cluster of Briese families. Further testing of individuals originating from Meseritz is needed to verify this. The Deutsch Krone Brieses could originate from Meseritz, or both clusters might originate from a common ancestor who lived in another region. Again more samples are needed to explore these possibilities.
With regard to the unrelated individuals, there are two possibilities; 1) they are descended from ancestors who independently took on the name Briese when surnames were being adopted, which was probably during the 15th century in eastern Prussia19 or 2) the surname lineage and genetic lineage have become disjunct at some stage during the generations elapsed since that time (e.g. in the case of adoption). In the latter case, King & Jobling (2009)20 reported that the rate of such events for English surnames was 2% per generation. This gives a 26% probability of a major genetic shift in a surname lineage in the 15 or so generations since the name was adopted, which is close to the 25% (2/8) of Briese individuals that were genetically unrelated to the others. That said, this does not disprove the possibility of multiple independent origins of the name and, in either case, these individuals are part of distinct lineages of the Briese surname. More samples might provide evidence whether or not these have also developed into distinct clusters of related families.
These results demonstrate that genetic genealogy is able to clarify relationships between families where paper records are missing. Several families from the Deutsch Krone area, other than those mentioned here, emigrated to Australia16 and North America5 and Y-DNA comparisons of their members should show how they are related. Broader sampling of Briese families from other areas should also be able to show whether the different clusters have originated independently or not. In the latter case, it should be able to show the patterns in related family clusters of a diverging lineage from a distant common ancestor.
More sampling is needed to clarify the relationships within family clusters and also the broader patterns of kinship between Briese families originating from different geographic regions. Based on the preliminary findings of this study, this should include:
1) broad-based sampling to discover whether there are more lineages to the Briese surname than the three already identified,
2) a mix of broad-based and targeted sampling to determine whether family-clusters exist within the two currently-isolated lineages and whether their geographic origins can be determined, and
3) targeted sampling to refine and add to kinship patterns amongst known families of the third lineage comprising Meseritz and the Deutsch Krone cluster.
Acknowledgements
I would like to thank the participants in the Briese surname project, not only for providing DNA samples but for genealogical information about their respective lineages. Thanks are also due to Gail Tonnesen, who started the Briese surname project and transcribed the records of the Lüben Evangelical Parish from microfilms held by the Family History Library in Salt Lake City, Utah. I would also like to thank the anonymous referees who provided useful insights into improving this paper. This article was peer reviewed with 5 commentaries.References
- Briese DT (2008), Why is the family tree a silver birch? The meaning of the Briese name
http://gang-gang.net/briesegenealogy/histories/briesename.php - Family Search (2014) Collection Records for German Birth, Deaths and Baptisms 1558-1898, German Marriages 1558-1929, German Deaths and Burials 1582-1958, Germany, Prussia, Brandenburg and Posen, Church Book Duplicates 1794-1874
https://familysearch.org/search/record/results?count=20&query=%2Bsurname%3ABRIESE&collection_id=(1473000%201473009%201491272%201494474 - Family Search (2014) Collection Records for User Submitted Genealogies - Briese.
https://familysearch.org/search/tree/results?count=20&query=%2Bsurname%3ABRIESE&collection_id=(2%203) - Verein fur Computergenealogie (2014) GEDBAS database
http://gedbas.genealogy.net/ - Briese DT (2014) The Briese Diaspora: emigration of Brieses from Prussia to the New World during the 19th Century.
http://www.gang-gang.net/briesegenealogy/histories/briese-diaspora.php - Tonnesen G, Briese DT (2011), Briese [DNA] Surname Project
http://www.familytreedna.com/public/briese/default.aspx - FTDNA is an abbreviation for Gene By Gene, Ltd doing business as Family Tree DNA, a commercial provider of genealogical DNA testing in Houston, Texas.
http://www.familytreedna.com/public/briese/default.aspx - McGee D (2008) Y-DNA Comparison Utility [web-based software]
http://www.mymcgee.com/tools/yutility.html - Bandelt, H. J., Forster, P., Rohl, A.. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16(1):37-48. DOI:10.1093/oxfordjournals.molbev.a026036.
http://dx.doi.org/10.1093/oxfordjournals.molbev.a026036 - Felsenstein, J. (2005) PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle.
http://evolution.genetics.washington.edu/phylip.html - Tamura K, Stecher G, Peterson D, Filipski A, and Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30: 2725-2729.
http://www.megasoftware.net/ - Genealogical Society of Utah (1987) Evangelische Kirche Lüben (Kr. Deutsch Krone) Kirchenbuch, 1757-1837 and 1860-1874 (microfilm)
- Canada RA (2013), If two men share a surname, how should the genetic distance at 37 Y-Chromosome STR markers be interpreted? FTDNA
http://www.familytreedna.com/learn/y-dna-testing/y-str/two-men-share-surname-genetic-distance-37-y-chromosome-str-markers-interpreted/ - Genealogical Society of Utah (1994) Evangelische Kirche Schwarzenau (Kr. Witkowo) Kirchenbuch, 1780-1903 (microfilm)
- Bielecki L (2013) Poznan Region Marriage Indexing Project for 1800-1899.
http://poznan-project.psnc.pl/ - Briese DT (2014) Briese Family Emigration to Australia
http://www.gang-gang.net/briesegenealogy/histories/briese-australia.php - Public Profiler World Names (2014)
http://worldnames.publicprofiler.org/ - Family Tree DNA Y Search database (2014)
http://www.ysearch.org/ - Immel, Uta-Dorothee, Krawczak, Michael, Udolph, Jürgen, Richter, Angela, Rodig, Heike, Kleiber, Manfred, Klintschar, Michael. Y-chromosomal STR haplotype analysis reveals surname-associated strata in the East-German population. European Journal of Human Genetics. 2006;14(5):577-582. DOI:10.1038/sj.ejhg.5201572.
http://dx.doi.org/10.1038/sj.ejhg.5201572 - King, T. E., Jobling, M. A.. Founders, Drift, and Infidelity: The Relationship between Y Chromosome Diversity and Patrilineal Surnames. Molecular Biology and Evolution. 2009;26(5):1093-1102. DOI:10.1093/molbev/msp022.
http://dx.doi.org/10.1093/molbev/msp022 - Fluxus (software)
http://www.fluxus-engineering.com