Here is a list of references, resources, and additional information relating to the presentation for Don’t Discard the Y-STRs:
Web Resources
- View the presentation slides online (PDF) (Updated 9 June 2017)
- Video Recording of live presentation available until 10 July 2017. Note that technical difficulties inhibited slide presentation.
- Genetic Homeland Tools
- Join a DNA Mapping Project
- DNA Marker Index
- Legacy Y-STR Marker Conversion Formulas
- NextGenMatch Tool (Requires free user account) Enables ad hoc comparison of any combination of SNP or STR markers). Good for Next-Gen results with inconsistent marker observations between two or more samples.
- International Society of Genetic Genealogy (ISOGG) Y-DNA Haplogroup Tree
- Dean McGee Y-Utility: Y-DNA Comparison Utility
- The Big Tree, YTree.Net by Alex Williamson
- YFull SNP List
- Integrative Genomics Viewer (IGV) tool for interactive exploration of large, integrated genomic datasets
- SAM Tools by Genome Research Limited a suite of programs for interacting with high-throughput sequencing data.
DNA Testing Laboratories for Genetic Genealogy of Y-Chromosome
Testing companies which offer both STR and SNP tests as Distinct Products.
- Gene By Gene, Ltd. doing business as Family Tree DNA (aka FTDNA)
- Y-Seq (Alpha Panel + Beta Panel + DYS442 has 37 STR markers)
Testing companies which do full genome (including Y-chromosome Next-Gen sequencing) by which SNPs and STRs may be determined from raw data with 3rd party tools.
Academic References and Other Sources
- Robert Sanders (2011) Are cancers newly evolved species? , UC Berkeley, Berkeley News
- Whit Athey (2007) Editorial on Kerchner’s Y-STR Haplotype Observed Mutation Rates in Surname Projects Study and Log, Journal of Genetic Genealogy 3(2)i-iii.
- Adamov et al (2015) Defining a New Rate Constant for Y-Chromosome SNPs based on Full Sequencing Data, ResearchGate from The Russian Journal of Genetic Genealogy Vol 7 No 1
- Henn et al (2009) Characterizing the Time Dependency of Human Mitochondrial DNA Mutation Rate Estimates. Mol Biol Evol 2009; 26 (1): 217-230. doi: 10.1093/molbev/msn244
- Campbell et al (2012) Estimating human mutation rate using autozygosity in a founder population. Nature Genetics. doi: 10.1038/ng.2418
- Taylor Edwards (2006) presentation “Laboratory Procedures, 2006” at the 2nd International Conference on Genetic Genealogy
- Yang et al (2014) Application of Next-generation Sequencing Technology in Forensic Science, DOI 10.1016/j.gpb.2014.09.001
- FTDNA (2014) Introduction to the BigY White Paper
- Zavodna et al (2014) The Accuracy, Feasibility and Challenges of Sequencing Short Tandem Repeats Using Next-Generation Sequencing Platforms, PLOS One 9(12): e113862 DOI 10.1371/journal.pone.0113862
- Darby et al (2016), Digital fragment analysis of short tandem repeats by high-throughput amplicon sequencing. DOI 10.1002/ece3.2221
- Payseur and Cutter (2006) Integrating patterns of polymorphisms at SNPs and STRs
- David Wilson et al R-M222 and Subclades DNA Project
- Brad Larkin (2016) Larkin DNA Project 2016 Update
- David Wilson et al (2017) R-M222 and Subclades DNA Project
- Brad Larkin (2017) Irish Mapping DNA Project
- Illustration – Human Genome – 23 Pairs of Chromosomes + mtDNA illustration in Butler JM (2005) Forensics DNA Typing 2nd Edition Figure 2.3, Elsevier Science Academic Press
- Illustration – DNA Replication by Mariana Ruiz on WikiMedia Commons. Note that while some of the finer details of the process may not be accurate in the diagram, the image conveys the idea that the double-helix structure of DNA is subject to mutations during replication or meiosis.
- James T. Robinson et al (2011) Integrative Genomics Viewer. Nature Biotechnology 29, 24–26
- Helga Thorvaldsdóttir et al (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics 14, 178-192
Illustrations of Trends and Findings in Y-Chromosome Genetic Genealogy in 2017
STR Samples by Year
Based on year of sample submission and whether it has been tested for Y-STRs.
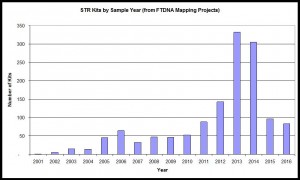
Trend illustrated with data from combination of Irish Mapping, English Mapping, and Scottish Mapping DNA Projects, 2017. n=1,369
Number of STR Markers Tested
Pie chart illustrating the proportion of each Y-STR panel which have been ordered for participants in the Irish Mapping, English Mapping, and Scottish Mapping DNA Projects.
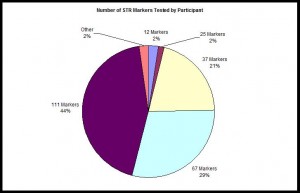
Pie chart illustrating the proportion of each Y-STR panel which have been ordered for participants in the Irish Mapping, English Mapping, and Scottish Mapping DNA Projects as of April 2017, n=1,384.
Histogram of STR Variance
Example within those confirmed with M222 positive SNP and how their variance at STR marker DYS390 using data from R-M222 and Subclades DNA Project.
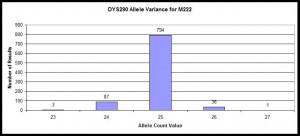
Histogram illustrating distribution of allele values for DYS390 for those confirmed with M222 positive SNP. Data from R-M222 and Subclades DNA Project, n=921
Histogram of SNP Subclades
Example within those confirmed with M222 positive SNP with histogram showing distribution of those lineages at Y-SNP markers classified as immediately below M222 on the current Y-chromosome phylogenetic tree.
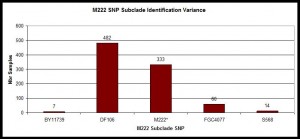
Histogram illustrating distribution of subclades of R-M222. Data from R-M222 and Subclades DNA Project, n=896