On April 20, 2018, Family Tree DNA (formally Gene By Gene Ltd, aka FTDNA) announced the initial release of allele count values for 389 new STR markers (labeled as Panel 6) scored from Next-Gen sequencing data recorded on Big Y DNA existing orders. Going forward FTDNA states they will only sell Next-Gen sequencing of the Y-chromosome as a single Big Y-500 product which will include Sanger sequencing of the current set of Y-111 markers as well as the Next-Gen sequencing for SNPs and STRs. (For background information on the difference between STR and SNP markers, Next-Gen technology, etc, see the Don’t Discard the Y-STRs presentation.)
In recognition of the importance of combining analysis of STR and SNP markers for genetic genealogy, FTDNA will insist on the Y-111 STR test being part of all future Big Y purchases:
Moving forward, customers who purchase a Big Y-500 will also have to purchase or upgrade to the Y-DNA111 test. However, customers do not have to have a Y-STR test already before purchasing the Big Y-500 since it contains the Y-111. Big Y without STRs will no longer be available.
Goal: Increased Phylogenetic Resolution and Clarity
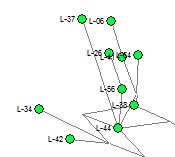
Illustration of Ambiguous Phylogeny within Larkin Type 01 from Y-37 STRs (2009) Homoplasy among Y-37 STR markers leads to phylogenetic tree construction with multiple mutation paths between individuals (e.g. L-42 to L-44 in the image).
For genetic genealogists, it is hoped that these new STRs will clarify the genealogical relationship within families and groups of men with similar Y-DNA markers. In many cases, the phylogenetic trees constructed from existing STR are ambiguous due to homoplasy or convergence at individual STR markers. The image at right shows part of a phylogenetic tree construction using Y-37 results. Where the mutational steps could be over different paths, there are multiple gray lines of parsimony between the individuals. So one goal for using the 389 new STR markers is to resolve the phylogeny and genealogical relationships among participants.
In order to assess the value of these Big Y-500 results, a quick study was made using eighteen (n=18) samples in the Larkin DNA Project who have Big Y-500 results and whose earliest known patrilineal ancestor used the Larkin surname (aka Larkin Ancestry Set). All the samples in this Larkin Ancestry Set are part of the R1b haplogroup.
Accounting for Unobserved Markers
Unlike the Sanger sequencing techniques used for FTDNA Y-12, Y-37, Y-67, and Y-111 products, the Next-Gen sequencing technology used in the Big Y production may not yield results at some positions / portions of the Y-chromosome. When the results for a particular marker are not reported, we refer to it as an Unobserved marker. The histogram below shows the frequency of Unobserved STR results among the new STR markers within results for the Larkin Ancestry Set. The median number of Unobserved markers was six (6) markers per sample with a range of 0 to 22 markers across all participants.
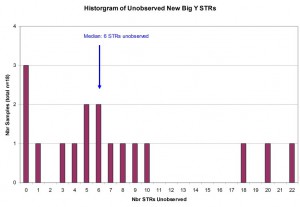
For new Panel 6 STR markers reported through Big Y-500 Next-Gen sequencing, here is the distribution of unobserved or unreported STR markers for Larkin Ancestry Set (n=18).
Genetic Distance between Larkin DNA Project Participants measured by Y-37 versus Big Y-500
An analysis was made of the percentage of STR markers matching and non-matching between all members of the Larkin Ancestry Set. In this pairwise analysis, a match or mismatch is only counted if both the individuals compared had an observed allele value at a particular marker (aka co-observed STRs).
e.g. Comparing Project participants L-0006 and L-0064 with Y-37 showed three markers different (34/37 matching = 91.9% match); with Big Y-500 these two individuals had four markers different across 492 markers observed in both samples (488/492 = 99.2% matching).
The graph below compares the percentage of marker matches from Y-37 versus Big Y-500 results for all the Larkin Ancestry Set compared to sample L-0006 as a reference. If the two types of test were equivalent, we would expect the results to fall roughly on the black diagonal line from lower left to top right. What we observe from the Big Y-500 (blue points and line) is that the percentage of matching markers is generally much higher for Big Y-500 results compared to the Y-37 markers. This finding implies that the new STRs in the Big Y-500 have a lot less variance than the original Y-37 marker set. In the most extreme difference, comparison of L-0006 and L-0040 showed only 46% matching on Y-37 but 92% matching on Big Y-500.
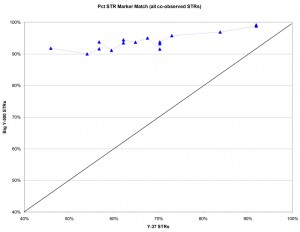
Larkin DNA Project participants who have both Y-37 and Big Y-500 STR test results were compared based on the percent of markers matching a reference participant.
Change in TMRCA Estimates within Larkin Type 01 Based on Big Y-500 STR Results
Four (4) of the Larkin Ancestry Set samples are part of the Larkin Type 01 subset – positive for the M222 SNP marker and the largest lineage for this surname. As illustrated earlier, there is evidence of homoplasy or backwards mutations in 37 and 67 STR marker tests making estimation of the patrilineal time to the most recent common ancestor (TMRCA) difficult. Expanding the number of STR markers used for estimation should enhance the resolution of the TMRCA estimate between individuals within Type 01. Additionally, some of the individuals within Type 01 have a known genealogical relationship. Being able to directly compare the mutational differences to the known genealogical relationship provides a means to directly calculate the STR mutation rate across the Big Y-500 markers. The graph below compares the TMRCA estimate between the L-0006 reference participant and the other three Larkin Type 01 participants using Y-37 and Big Y-500. For this group at least, the Big Y-500 observed mutation rate implies that there is about 40 years in TMRCA for each marker difference between two participants using Big Y-500 STRs.
Also note that in two of the three comparisons, the Big Y-500 results significantly reduced the TMRCA estimates, suggesting these participants are more closely related than the Y-37 test had indicated. This concept is generally consistent with the findings of upgrading earlier generations of Y-12, Y-25, and Y-37 tests: two persons that are truly related genealogically will see their genetic distance (mismatches) be stable when additional markers are tested. Two persons who are not genealogically related will typically see their genetic distance grow dramatically as more markers are added for comparison.
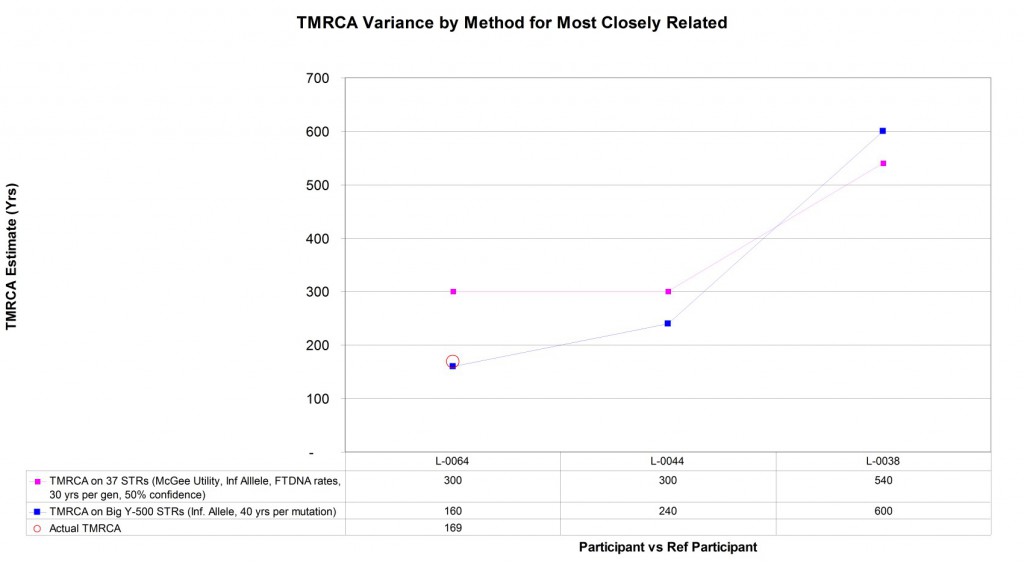
Graphic comparison of patrilineal time to most recent common ancestor (TRMCA) estimates drawn from Y-37 and Big Y-500 STR marker mutations within Larkin Type 01 group.
Summary
In summary, the new STRs from Big Y-500 do help refine phylogenetic and TMRCA estimates over the genealogical time frame. As with earlier generations of genetic genealogy testing, interpretation of STR matches should try to incorporate comparison of SNPs for a full understanding of relatedness based on the Y chromosome. With continued refinement of the SNP phylogenetic tree thanks to the growth of the Big Y database, the importance of having these additional STRs for distinguishing within kin groups will be increasingly clear.